The Forensic science is the collection of disciplines that scientifically contribute to the legal system; for instance, pathology, Odontology, anthropology, chemistry, toxicology, and genetics. Forensic genetics is the area in forensic science where DNA analysis is used for molecular identification of biological material found at crime scenes.
DNA analysis is now one of the most definitive techniques of identification in “forensic science”, paternity testing and missing individuals. It is a very effective technique of human identification.
Short tandem repeats (STRs) are commonly utilized as DNA markers in paternity testing and criminal investigations due to their high genetic variation among individuals in population. X-chromosome short tandem repeat (X-STR) markers are a significant addition to paternity and forensic casework. They've been beneficial in deficient paternity testing when the mother is accessible for typing. Both males and females retain one of their mother’s X chromosomes, and females retain their second X chromosome from their father. So, female individuals fathered by the same man share their paternal X chromosome and the other one X chromosome is the same with the mother. Hence, in case of deficiency paternity in which the mother is available for typing, the possible X alleles of the putative father can be determined and the paternal profile can be reconstructed. Deficiency paternity cases, characterized by the absence of the alleged father, are a challenge for forensic genetics. Furthermore, there are cases that show the effect of additional X-STR markers in identifying cases that cannot be solved using autosomal markers (e.g., special reverse paternity cases).
There were 33 X-STR loci that have been used within the forensic community. As with autosomal STR loci used in forensic analysis, tetranucleotide repeats are most commonly selected due to lower stutter product formation compared to dinucleotide or trinucleotide repeats.
The X-STR is a complementary tool to autosomal STR, (Y-STR) and (mtDNA) markers. It can be used in forensic investigations like complex kinship analysis. DNA testing of X-chromosomal STR (X-STR) polymorphisms has been the main focus in a number of researches, primarily due to its applicability in the analysis of population genetic research by using multiplex polymerase chain reactions (PCR) for use in DNA testing in forensic application. In other words, multiplex PCR is widely used for the study of population genetics, and as well as forensic.
Methods
Study population
The population of this study includes 200 males apparently healthy unrelated participants from different region of Baghdad City, their ages ranged between 20 and 50 with mean age (36.83 ± 7.2) years. This study was conducted in a College of Biotechnology at Al-Nahrain University during the period from January 2019 to April 2020. Each participant was asked a systematic questionnaire for the various etiological factors of genetic disease, history of parents, relatives, and X-linked diseases such as hemophilia, G6PD dehydrogenase deficiency, and color blindness. This study was approved by the congress at the College of Biotechnology, Al-Nahrain University. Ethical consideration written consent was obtained from all participants and the researcher explained the objective of the study, signed written consent was taken from each individual participating in the study.
Extraction of genetic material
Two milliliter venous blood samples were collected from participants into an EDTA tube for DNA extraction; sample was stored at − 23 °C until use. Total genomic DNA was extracted from frozen blood using the WIZPREP™ DNA Extraction Kit supplied by (Korea).The PCR product was sent for to Macrogen Company (Korea), and then the PCR product size was compared with the information about X-STR reference allele and size. The microsatellite analysis was performed by Geneious Prime software.
Power of discrimination (PD)
The probability that two randomly selected individuals will have different genotypes. Power of inclusion (Pi): sum of the squares of expected genotype frequencies. The following formula for calculating the power of discrimination in male according to.
Polymorphism information content (PIC)
Refer to the value of a marker for detecting polymorphism within a population. The PIC using following formula according to. It was used allelic frequencies marker.
Statistical analysis
The Statistical Analysis System (SAS) (2012) program was used to detect the effect of different factors in study parameters.
Results
Amplification of X-STR
After optimization of primers, the multiplex PCR was done for each three primers set at 58 °C, using hot start Promega master mix as shown in Fig. 1.
Allele distribution and frequencies
The high, low, and rare allele
In the current study, twelve X-STR loci were grouped into four groups based on their molecular size (base pair). The first group includes DXS7424, HPRTB, and DXS8377. Second group involves GATA31E08, DXS7423, and DXS8378. The third group consists of DXS9895, DXS10074, and DXS6809. Fourth group includes DXS7133, DXS101, and DXS6807.
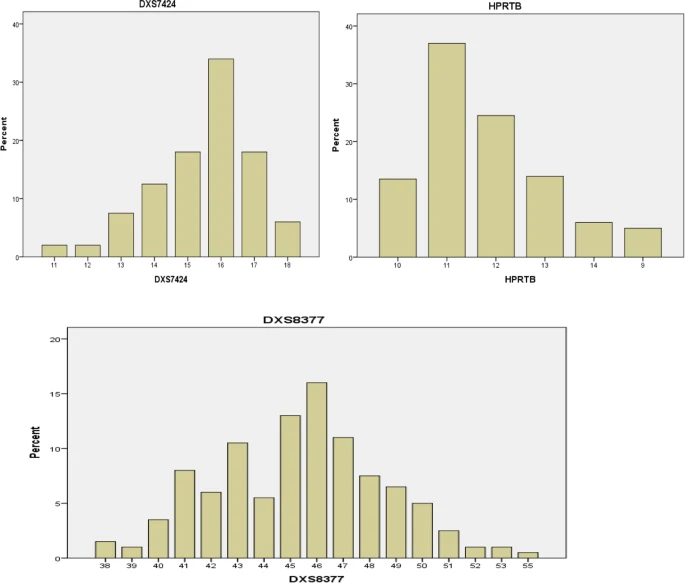
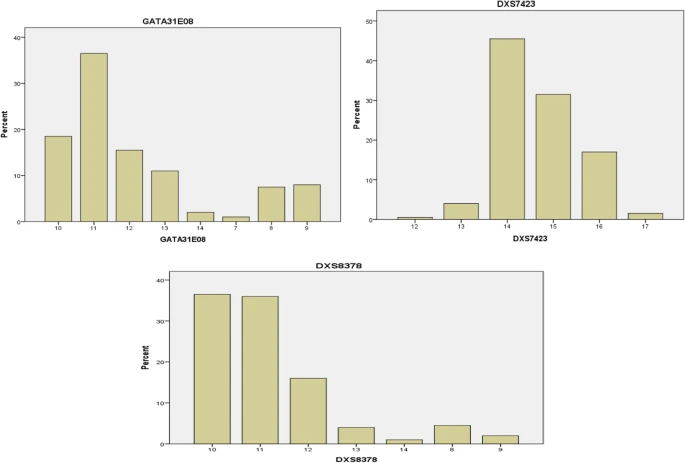
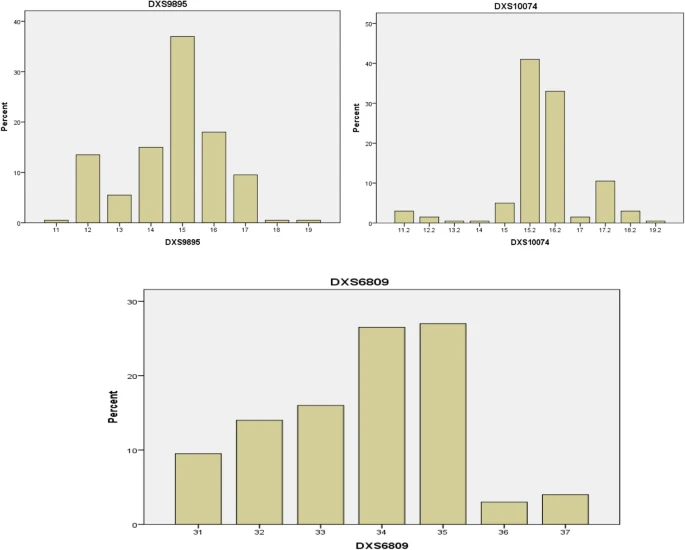
The distributions of the observed alleles and genotype frequencies for the 200 unrelated Arabic Iraqi males was shown in Table 3 and Figs. 2, 3, 4, and 5. The data from 200 unrelated Arabic Iraqi males was transformed to allele occurrence by counting the number of times of each allele that was identified and the results listed in Table 4.
Discussion
Over the last decade, the usage of X chromosomal short tandem repeat (STR) markers has increased dramatically in the forensic field. This current study was conducted to analyze the genetic of (X-STR) frequencies in Arab Iraqi male population. The purpose of this study was to explore at the frequency of 12 (X-STR) haplotypes in the Iraqi population as a reference data source for individual identification. As allele frequency, the distributions of observed alleles and genotype frequencies for the 200 unrelated Arab Iraqi males were demonstrated. The results of the present study revealed the high and low frequency of these alleles. Our findings revealed that the DXS7424 locus of Iraqi males showed the range between 0.02 and 0.34 with high frequency for allele 16 and low frequency for alleles 11 and 12. Relative to the DXS7424 locus, previous study by Nakamura Y and Minaguchi K., 2010 showed that the alleles in Japanese population were 12, 13, 14, 15, 16, 17, and 18 and frequency ranged from 0.011 to 0.445 with similar high frequency (0.445) for allele 16.
According to the above results concerning HPRTB locus, the allele frequency ranged from 0.05 to 0.37 with high and low frequency for allele 11 and allele 9 respectively. The study by Hameed et al. 2015 conducted in Iraq found that the alleles in Iraqi population were 7, 8, 9, 10, 11, 12, 13, 14, 15, and 16 and frequency ranged from 0.002 to 0.436 with high frequency (0.436) for allele 13. This finding could be related to the use of argus 8X-STR specific X-STR kits, whereas in this study, the authors employed multiplex PCR from 12 loci as described by Nakamura Y and Minaguchi K ., 2010.
In the DXS8377 locus, the high, low, and rare frequency alleles were for allele 46, alleles 52, 53, and allele 55 respectively with frequencies ranged from 0.05 to 0.16. However, the findings by Poetsch et al. 2005 discovered that the alleles at this locus were 39, 40, 41, 42, 43, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, and 56 in German population and the allele 51 had a high-frequency percentage (0.138).
Regarding to the GATA31E08 locus, the high and low allele frequency alleles were observed in allele 11 and 7 respectively and the allele frequency ranged from 0.01 to 0.365. It was found by Nakamura Y and Minaguchi K., 2010 that the most prevalent alleles in the Japanese population were 7, 8, 9, 10, 11, 12, 13, and 14, with a frequency ranging from 0.001 to 0.307, with allele 11 having the highest frequency (0.307).
In the present study, the DXS7423 locus had a high and low allele frequency showed in allele 14 and 17 with a rare allele showed in allele 12. Similarly, previous studies done by Al-Snan et al. 2019 and Hameed et al. 2015 reported that the allele 14 at DXS7423 locus emerged as a high-frequency allele with allele frequency 0.462 and 0.409, and this locus exhibit low polymorphic alleles. The study done by Nakamura Y and Minaguchi K., 2010 showed that the alleles in Japanese population was 13,14,15,16 with frequency ranged from 0.005 to 0.608 with high frequency (0.608) for allele 15.
As a result, the DXS8378 locus had a high and low allele frequency for allele 10 and allele 14 with a range from 0.01 to 0.365. In a contrast with the present study, previous studies in Iraq and Germany revealed that the allele 11 having the highest frequency (0.371 and 0.374) respectively.
The DXS9895 locus showed the high allele frequency for allele 15 and low frequency for allele 13 with a rare allele reported for allele 11, 18, and 19. Such observation suggested by Poetsch et al. 2005 upon the DXS9895 locus found that the allele 24 having a high frequency.
The allele frequencies at DXS10074 locus ranged from 0.005 to 0.41 and the allele 15.2 appeared in high frequency and allele 12.2 and 17 in low frequency and as a rare allele found in allele 13.2, 14, and 19.2. A previous study done in Iraqi and Turkish population revealed that the high frequency allele at the DXS10074 locus was 15.
The allele frequencies at DXS6809 locus ranged from 0.03 to 0.27, and the allele 35 appeared in a high frequency while the low frequency appeared in allele 36. Such a result of previous study in Japan revealed that the allele repeats in Japanese population was 26, 29, 30, 31, 32, 33, 34, 35, 36, and 37 and frequency range from 0.003 to 0.309 with a high frequency for allele 33.
The allele frequencies at DXS7133 locus ranged from 0.02 to 0.46 and the allele 11 appeared in a high frequency while the low frequency was found in alleles 15 and 16. The findings obtained by Nakamura Y and Minaguchi K., 2010 conducted in japan showed that the allele repeat in Japanese population was 9, 10, 11, and 12 and frequency range from 0.06 to 0.7 with a high frequency for allele 9.
The allele frequencies at DXS101 locus ranged from 0.015 to 0.22 and the allele 25 appeared in high frequency with low frequency observed in allele 22 and 29. The result of previous study in Germany showed that the allele repeat in German population was 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, and 30 and frequency range from 0.03 to 0.2 with high frequency for allele 24.
The allele frequencies at DXS6807 locus ranged from 0.005 to 0.405 and the allele 11 appeared in high frequency while the low allele frequency showed in allele 17 and allele 18 found as a rare allele. A prior investigation carried out in japan showed that the allele repeat in Japanese population was 11, 12, 13, 14, and 15 and frequency range from 0.08 to 0.370 with similar a high frequency for allele 11.
Rare alleles are polymorphic alleles that occur in less than 1% of the population. However, the frequency of detecting functional uncommon alleles was highly dependent on the sample size.
The power of discrimination (PD) is the chance that choosing two individuals randomly would not have matching DNA profile. The current study found the PD value ranged from 0.663 for DXS7423 locus and 0.9066 for DXS8377 locus. This result is close to Iraqi study that showed the DXS7423 locus had 0.522 of PD value and an Italian study the PD value was 0.913 for DXS8377 locus.
Polymorphism information content (PIC) is evaluated by the markers ability to identify polymorphism in the population based on the number of alleles detected and the frequency of their distribution. Therefore, PIC determines the marker’s discriminatory capability, basically depending on the number of recognized (identified) alleles and their frequency of distribution. The result of the present investigation revealed that the PIC for all twelve loci were appeared more than 0.5, indicating good informativeness of all X-STR markers.
In order for novel alleles to be noted in DNA profiles, consideration should be given to these new alleles for forensic science recommendations with a view to being included in the DNA database, and in order to assign frequency to estimate the probability of a specific DNA profile across a population of interest (random match probability).
The distribution occurrence is another measure to examine the frequencies of the most common occurrence genotypes in order to identify the usefulness for particular set of DNA markers would therefore be the least powerful in terms of being able to differentiate between two unrelated individual.
The X-STR loci become main completely and alternative uses for forensic application especially, kinship testing, miss distress, and more efficient use degraded DNA and is widely used in many complicated cases.
Conclusions
Our result found that in Iraq Arab population, the highest forensic efficiency parameter was DXS8377 locus that has the highest polymorphic allele. In contrast, the DXS7423, DXS9895, DXS10074, and DXS6807 loci have a rare allele with such have a little role in the Iraqi population database. The finding of the present study is relevant to major concerns in forensic science, such as population geneticists’ inquiry into the behavior of rare variants, with an emphasis on alleles with low relative frequency. As a recommendation in this study, we suggested to focus the level of X-STR loci polymorphism in a three major ethnic groups in the Iraq to start for building X-STR data base.